Wheat is the second most important grain crop produced in South Africa, playing a significant role in both cultural and economic advancement worldwide. Wheat is crucial for ensuring food security beyond national borders, as evidenced by recent volatility in wheat markets.
The relatively low production of wheat across Southern Africa is primarily due to a range of interacting biotic and abiotic stresses.Recent studies have demonstrated that beneficial microbiomes from the wheat rhizosphere can enhance the health and crop yield of plants cultivated in challenging environments.
From the field to the Lab
 Our Project includes the collection of rhizosphere soil samples from both an irrigated and a rain-feed Farm in the Western Cape, South Africa. The samples were collected at three growth stages of wheat and then transported to the laboratory for metagenomic and metabolomic studies.
Our Project includes the collection of rhizosphere soil samples from both an irrigated and a rain-feed Farm in the Western Cape, South Africa. The samples were collected at three growth stages of wheat and then transported to the laboratory for metagenomic and metabolomic studies.
Plant microbiome research increasingly uses high-throughput sequencing of marker gene amplicons and whole-genome sequencing from metagenomic samples to clarify the make-up, structure, and spatial distribution of microbial communities in the environment.
Modern sequencing technologies offer the advantage of being highly selective, capable of targeting specific groups of microorganisms (such as bacteria or archaea) or even specific functional genes present in the microbiota.
From Lab to Sequencing
 In the laboratory at the University of Pretoria, several processes, including microorganism isolation, physicochemical analysis, and DNA extraction, are conducted.
In the laboratory at the University of Pretoria, several processes, including microorganism isolation, physicochemical analysis, and DNA extraction, are conducted.
Once DNA is extracted from the rhizospheric soil, it is sent for long-read PacBio sequencing by the Inqaba Biotec sequencing provider (https://inqababiotec.co.za/).
Bioinformatic methods have proven effective in classifying microorganisms based on taxonomy, metabolism, habitat prediction, genome reconstruction, and structural variations. Long-read DNA sequencing methods (e.g., PacBio, Nanopore) are anticipated to improve the quality of genome- and metagenome-derived sequences and to overcome the limitations of short-read (e.g., Illumina) binning and assembly in highly complex samples.
Data analysis
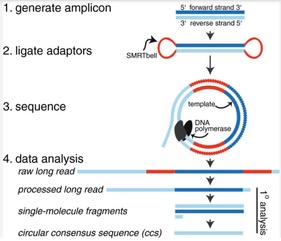 The raw reads obtained from sequencing are then analyzed using bioinformatic tools to:
The raw reads obtained from sequencing are then analyzed using bioinformatic tools to:
• Examine the microbial diversity associated with cultivated wheat plants.
• Study dynamic changes in the composition of wheat-associated microbiota.
• Select promising members of the natural wheat microbiome for agricultural application in South Africa.
Discovery highlights
The application of the latest SMRT PacBio technology in metagenomics has enabled the identification of key microbial species that exhibit overabundance and centrality within microbial communities associated with the wheat rhizosphere. These findings pave the way for potential applications in sustainable agriculture, such as development of microbial inoculants to enhance wheat resilience against environmental stressors and phytopathogens. Key finding of the study are as follows:
- The microbiome associated with the wheat rhizosphere in South Africa differs significantly from those identified by our collaborators in Italy and Spain, highlighting the necessity of creating biofertilizers and biocontrol agents specifically suited to the region of their application.
- The use of network-based methods in studying dynamic changes in rhizomicrobial composition during plant development and seasonable whether changes has enabled a deeper understanding of the complex interactions between microbial and fungal species in soil and plant environments.
- New approaches of utilizing advanced sequencing technologies for epigenetic profiling of metagenome-assembled contigs have been proposed to estimate the levels of abiotic stress affecting different community members.
What is next?
A collection of prospective microorganisms from the wheat rhizosphere was assembled for further studies on their applicability in tropical agriculture as soil fertilizers and biocontrol agents against plant pathogens.
Want to know more?
Contact catherine.malingreau@wagralim.be or Follow the project’s LinkedIn page

